Welcome Maya Weissman!
We are delighted to welcome postdoctoral scholar Maya Weissman to our group. She is joining us after having successfully defended her PhD on bet hedging at Brown University with Prof. Dan Weinreich. Welcome Maya!

We are delighted to welcome postdoctoral scholar Maya Weissman to our group. She is joining us after having successfully defended her PhD on bet hedging at Brown University with Prof. Dan Weinreich. Welcome Maya!

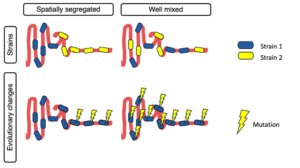
We are delighted to share our latest manuscript on the distribution of genetic diversity along the gut. In this work, we analyze the luminal contents of mice inoculated with the same human stool sample and find to our surprise that genetic diversity is uniform along the gut, rather than spatially segregated as observed at the species level.
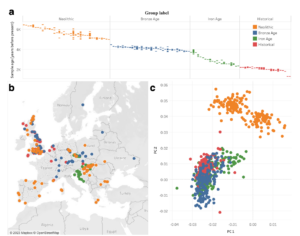
We are happy to share that our manuscript on Leveraging ancient DNA to uncover signals of natural selection in Europe lost due to admixture or drift is now officially out in Nature Communications. In this paper we perform a haplotype homozygosity selection scan in aDNA from Europe.
Congratulations to Peter for winning a departmental travel award, and to Ricky and Mariana for winning Dissertation Year Awards from UCLA!
We are excited to work with Alexandria Hunt and Evelyn Barajas this summer, both Bruins in Genomics students!
Alexandria is an undergraduate student at UCLA in Computational and Systems Biology, and Evelyn recently graduated from Heritage University in Toppenish, WA with a B.S.
in Computer Science.


Left: Evelyn, Right: Alexandria
Richard Wolff recently presented his paper at the Microbiome Virtual International Forum and received an award for best talk in his session!
See the talk here:
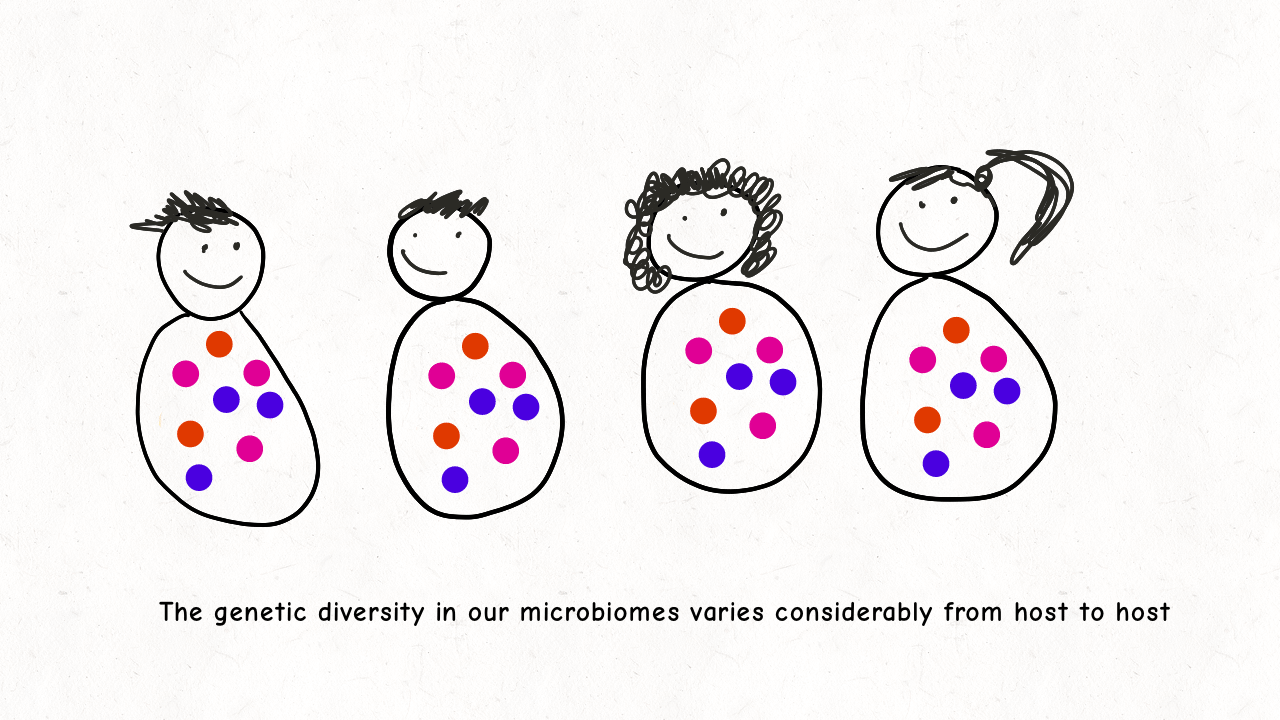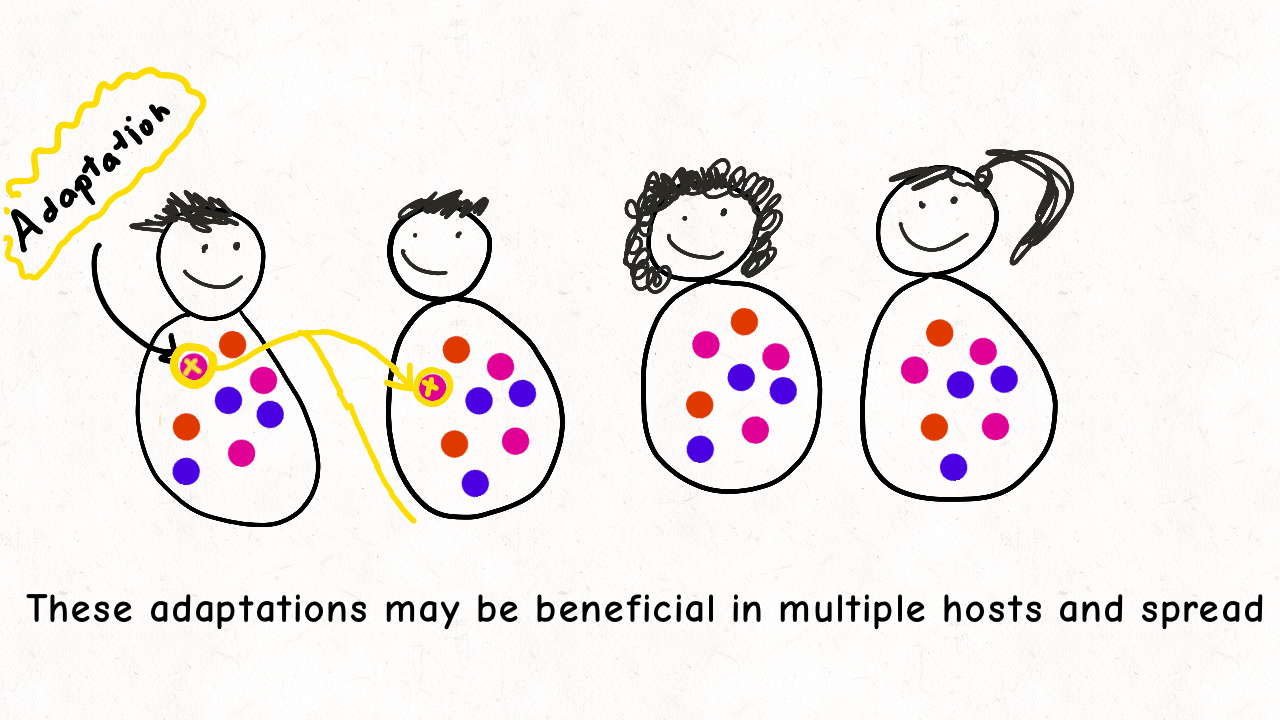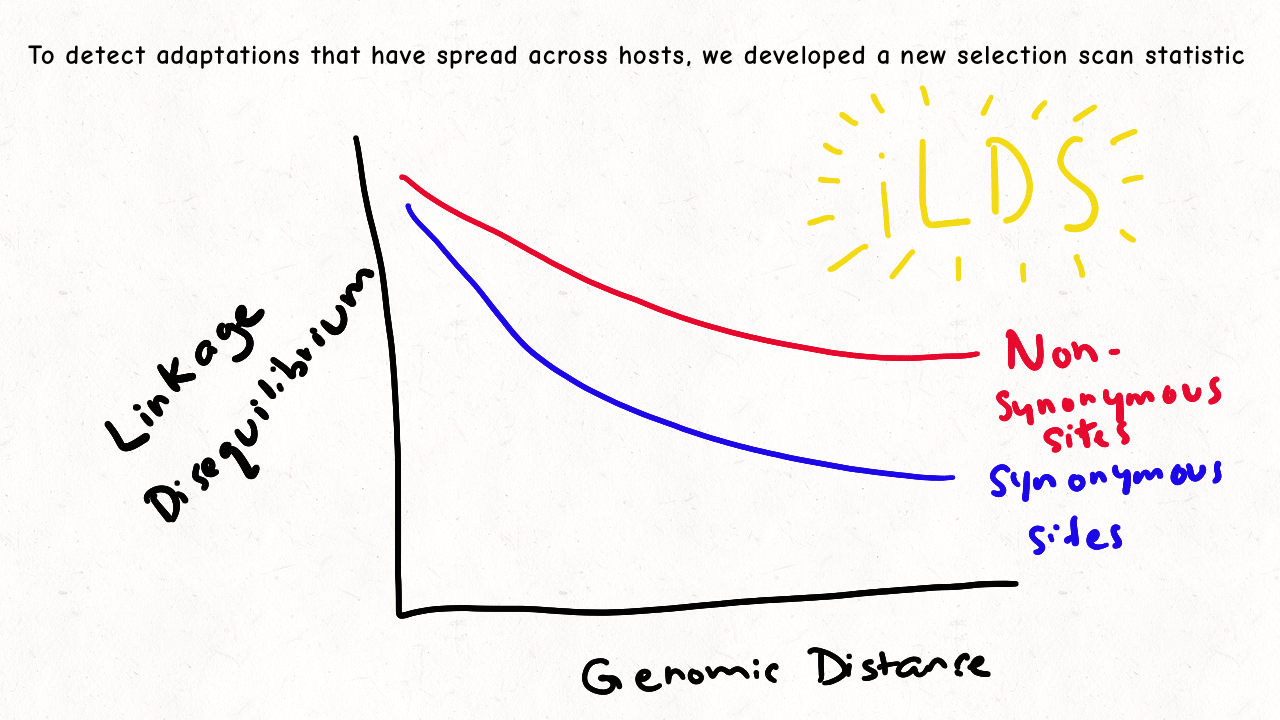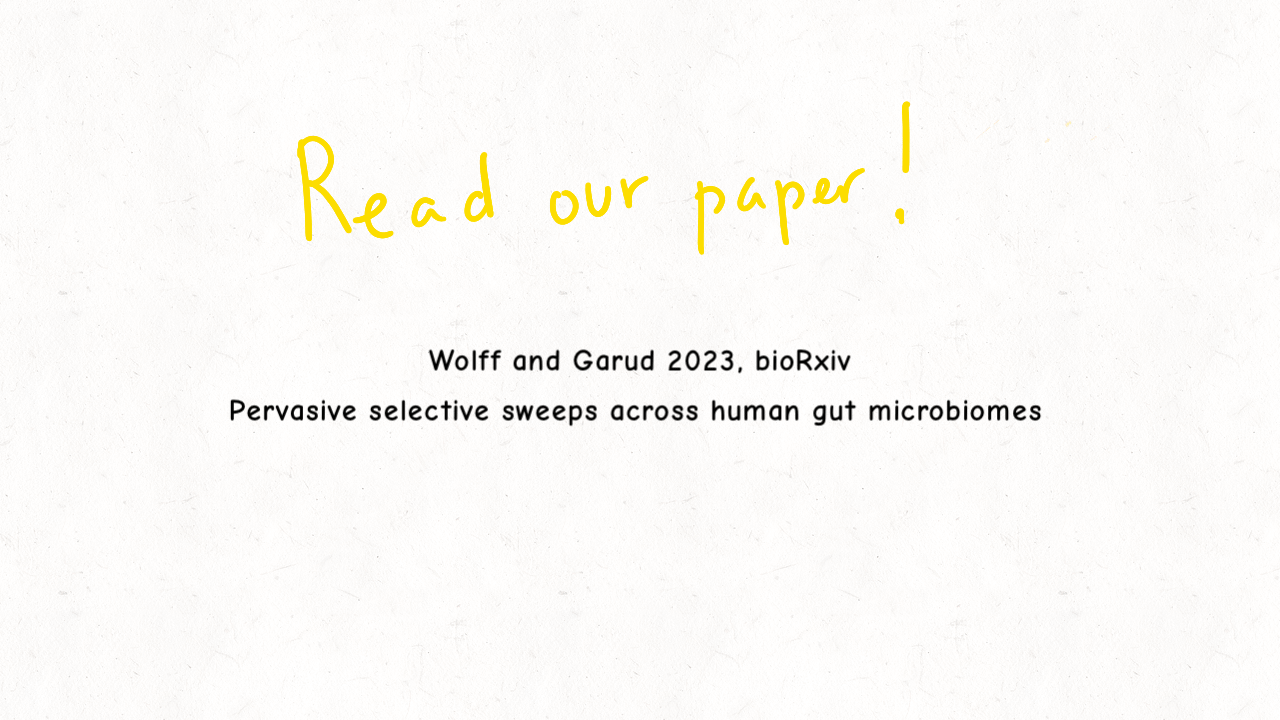
Richard Wolff and I are delighted to share our latest preprint entitled “Pervasive selective sweeps across human gut microbiomes.” Please check out our paper and our animation featuring the paper below!
We are so proud of Mariana Harris for winning the DeLill Nasser Award! She will be attending PEQG this Spring with the award to present her work on the prevalence of hard versus soft sweeps on the X chromosome of Drosophila.
See her publications in Genetics (2024) and MBE (2023)!
We are incredibly proud of doctoral student Michael Wasney for winning the inaugural UCLA Goodman-Luskin Microbiome Center seed fellowship!

We are delighted to share our latest preprint on “Inference of the demographic histories and selective effects of human gut commensal microbiota over the course of human history“. Please check out our paper and below is an animation we made sharing a few highlights from the paper!
