Evaluating supervised and unsupervised background noise correction in human gut microbiome data
We are thrilled to share that our paper on Evaluating supervised and unsupervised background noise correction in human gut microbiome data is finally out in PLoS Comp Bio! In this paper, we assess the ability of several noise correction methods to improve phenotype prediciton ability as well as biomarker discovery. We find utility in a Principal Component correction approach commonly used in other domains but to date has had seen limited application to microbiome data. Congrats to first-author Leah Briscoe!
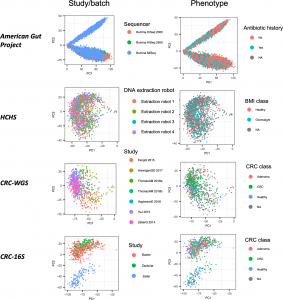
Figure 1 from our paper: Top PCs can sometimes correlate more with dataset label than disease. PCA applied to CLR-transformed taxonomic abundance data from the four datasets of the study. Each point represents a single microbiome sample colored by either study or batch and by phenotype group.