Population Genetics in the Microbiome — Review for Trends in Genetics
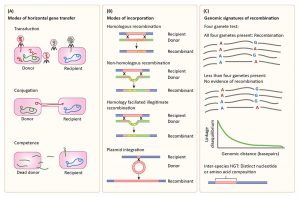
We are very happy that our review on Population Genetics in the Microbiome has been recently accepted to Trends in Genetics! The review will be officially published in January, but in the meanwhile, a copy is available here. Below is one of our figures illustrating modes of horizontal gene transfer, incorporation of DNA into the receipient’s genome, and statistical signatures of recombination.