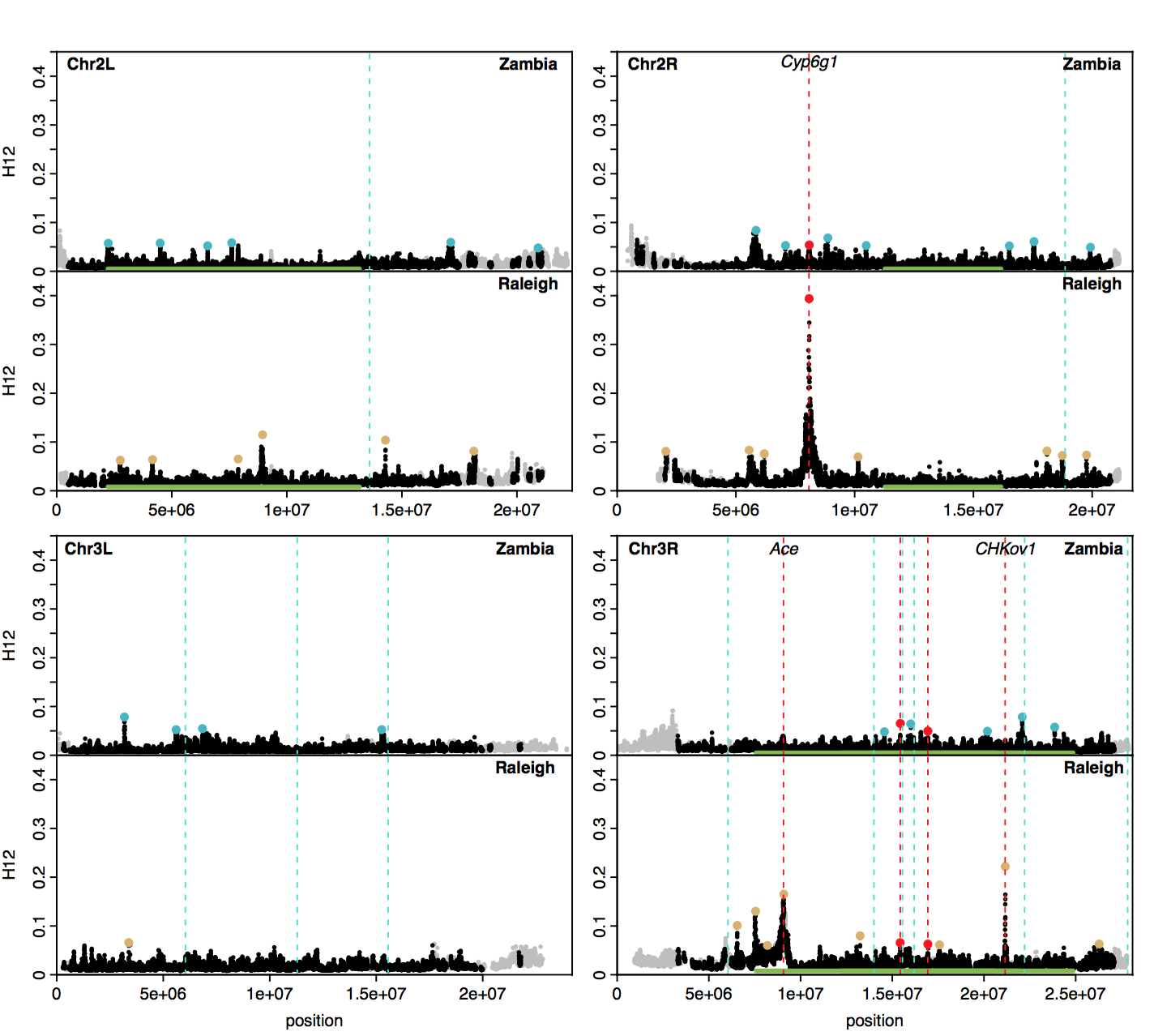
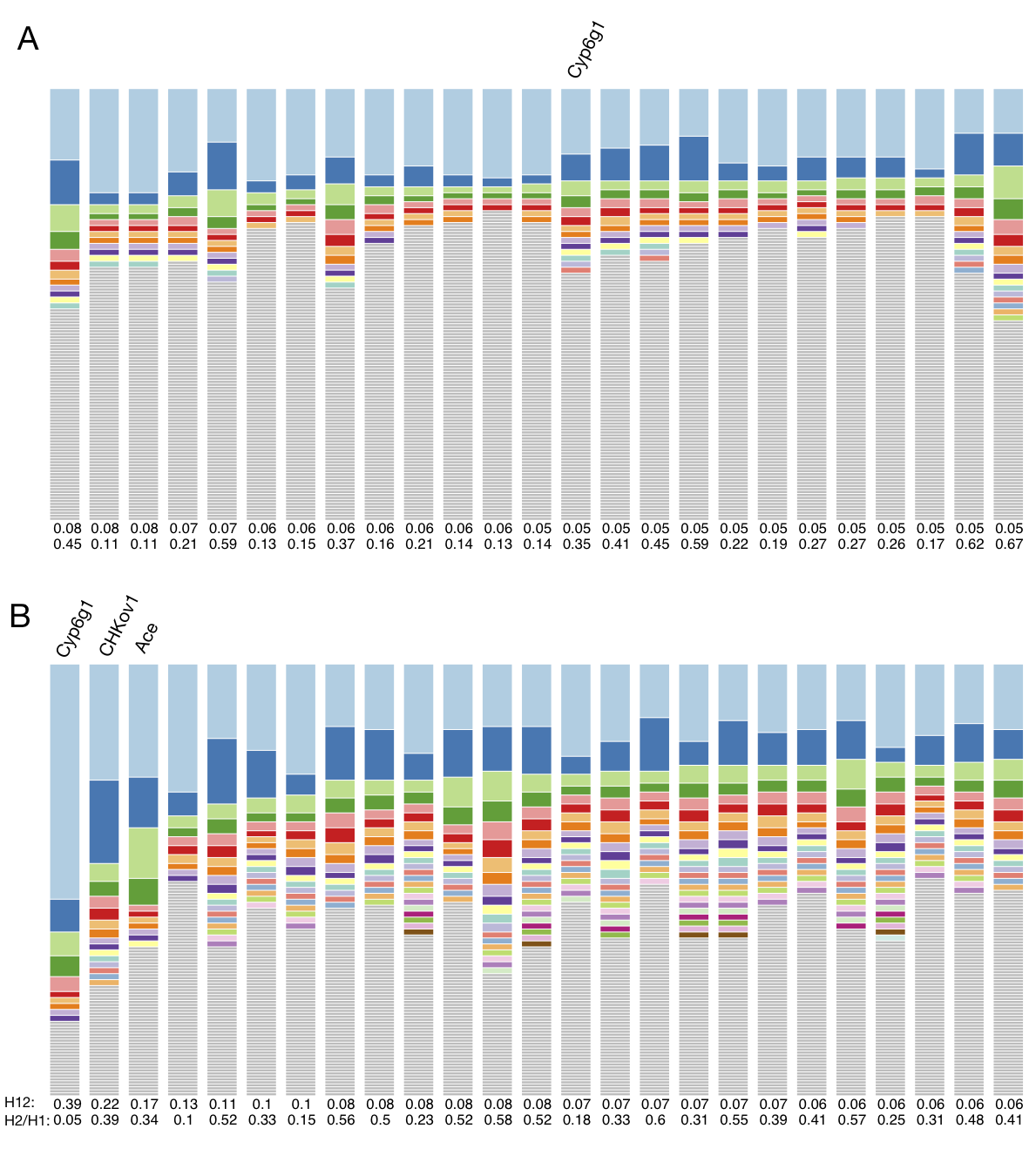
Do adaptive TEs overlap with top H12 peaks in Raleigh or Zambian data?
Recently, my colleague asked if adaptive TEs overlap with the top H12 peaks identified in Raleigh or Zambian D. melanogaster data. A paper by Gonzelez et al. 2010 shortlisted 13 adaptive TE candidates in D. melanogaster. One of the candidates is at the locus CHKoV1, which has been experimentally validated to confer resistance to pesticides.
Below I am replicating a figure from my paper showing an H12 scan in both Zambian and Raleigh D. melanogaster populations. The peaks highlighted with blue and beige dots indicate positions of the top 25 H12 peaks in the Zambian and Raleigh scans, respectively. I highlighted with red vertical lines the positions of three positive controls, Cyp6g1 (at this locus another TE confers DDT resistance, see this paper), Ace, and CHKoV1. Two additional red lines highlight peaks that are shared in both populations at unidentified loci. Additionally, I highlighted with blue vertical lines the positions of the candidates that Gonzalez et al. identified. Gonzalez et al. also identified candidates on the X chromosome, but we did not include an analysis of the X in our paper. As can be seen, the main overlap is at the CHKov1 locus. There are close overlaps at a peak on Chr2R in Raleigh and a couple peaks on Chr3R in Zambia, but it is difficult to know for sure if these are true overlaps given that the peaks are shifted slightly relative to the loci of the genes in which the TEs are located.