CTEG talk at Berkeley
Nandita recently gave a seminar at Berkeley and her colleague, Alison Feder, drew a graphical abstract of the talk!
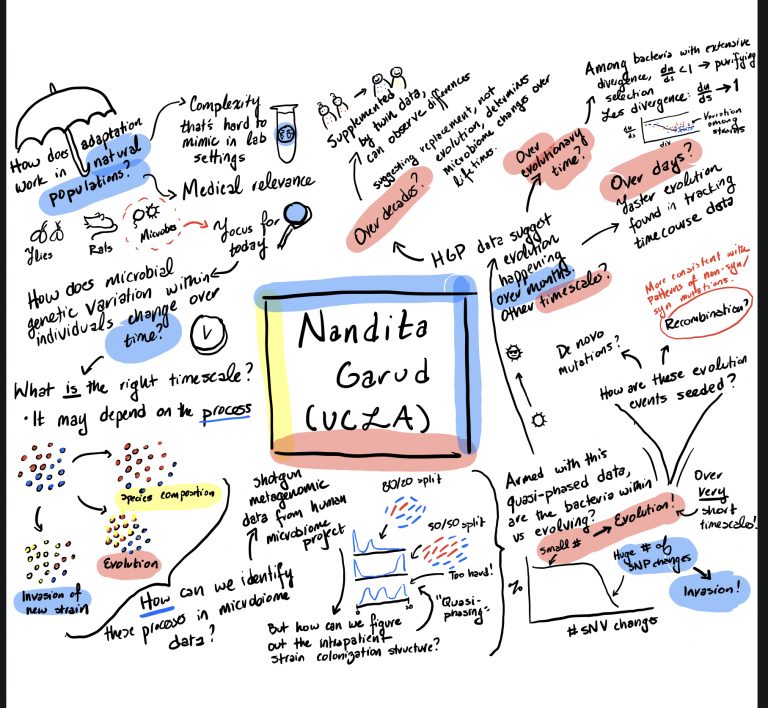
Nandita recently gave a seminar at Berkeley and her colleague, Alison Feder, drew a graphical abstract of the talk!

Nandita is a Microbiome, Neurobiology and Disease Scialog Fellow and recently participated in the annual Scialog conference to innovate and pitch new ideas about the connection between the gut and the brian.
This quarter, Prof. Nandita and TA Ricky are teaching a new R-based course on Evolutionary Genomics for undergraduates. Our goals for the class are to teach students how to make evolutionary inferences from DNA sequences. We are covering topics ranging from evolutionary dynamics on short time scales within populations to longer-term evolutionary forces across across species. We conclude by examining the microbiome. Here is our syllabus.
We are very proud of Mariana Harris for receiving the UCLA GATP fellowship and Ricky Wolff for receiving honorable mention in the NSF GRFP competition!! Congrats!
We are very proud to share our recent preprint:
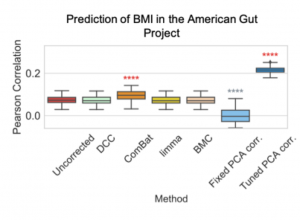
Correcting for background noise improves phenotype prediction from human gut microbiome
Leah Briscoe, Bruna Balliu, Sriram Sankararaman, Eran Halperin and Nandita Garud
Many technical and biological variables can contribute to variation in microbiome data. For example, in this combined dataset, the study contributes more variation than colorectal disease status.

Many of the popular methods for correcting background noise in microbiome data are supervised. Unsupervised methods may be preferable because they can correct for unmeasured sources of variation. We perform a comparative analysis of background noise correction methods and find that regressing out Principal Components after applying a centered log-ratio transformation is quite effective in removing unwanted variation. In fact, if we correct for background noise, we can even improve phenotype prediction of many phenotypes.
Nandita has been named an Allen Distinguished Investigator! The Garud lab is excited to be collaborating with Drs. Carolina Tropini, Aida Habtezion, and Siddharth Sinha on the following grant: Distinct immune-metabolic niches in inflammatory bowel disease. Link to press release.
We are delighted to share our latest preprint on:
Comparative Population Genetics in the Human Gut Microbiome
William Shoemaker, Daisy Chen, and Nandita Garud
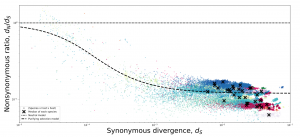
The genetic variation in the human gut microbiome is responsible for conferring a number of crucial phenotypes like the ability to digest food and metabolize drugs. Yet, our understanding of how this variation arises and is maintained remains relatively poor. Thus, the microbiome remains a largely untapped resource, as the large number of co-existing species in this microbiome presents a unique opportunity to compare and contrast evolutionary processes across species to identify universal trends and deviations. Here we outline features of the human gut microbiome that, while not unique in isolation, as an assemblage make it a system with unparalleled potential for comparative population genomics studies. We consciously take a broad view of comparative population genetics, emphasizing how sampling a large number of species allows researchers to identify universal evolutionary dynamics in addition to new genes, which can then be leveraged to identify exceptional species that deviate from general patterns. To highlight the potential power of comparative population genetics in the microbiome, we re-analyzed patterns of purifying selection across ~40 prevalent species in the human gut microbiome to identify intriguing trends which highlight functional categories in the microbiome that may be under more or less constraint.
We are delighted to be working with rotation students Helen Huang, Nicole Zeltser, Jon Mah, Alejandro Espinoza, and Albert Xue this year!
This summer we have a full house with many interns joining us, as well as new incoming PhD students including Albert Xue, Jon Mah, and Mariana Harris! Here’s our group meeting over Zoom!
This summer we are delighted to welcome four new BIG (Bruins in Genomics) summer interns to our group: Shavonna Jackson, Etan Dieppa, Kevin Delao, and Maya Singh!

Etan Dieppa

Maya Singh

Shavonna Jackson

Kevin Delao
